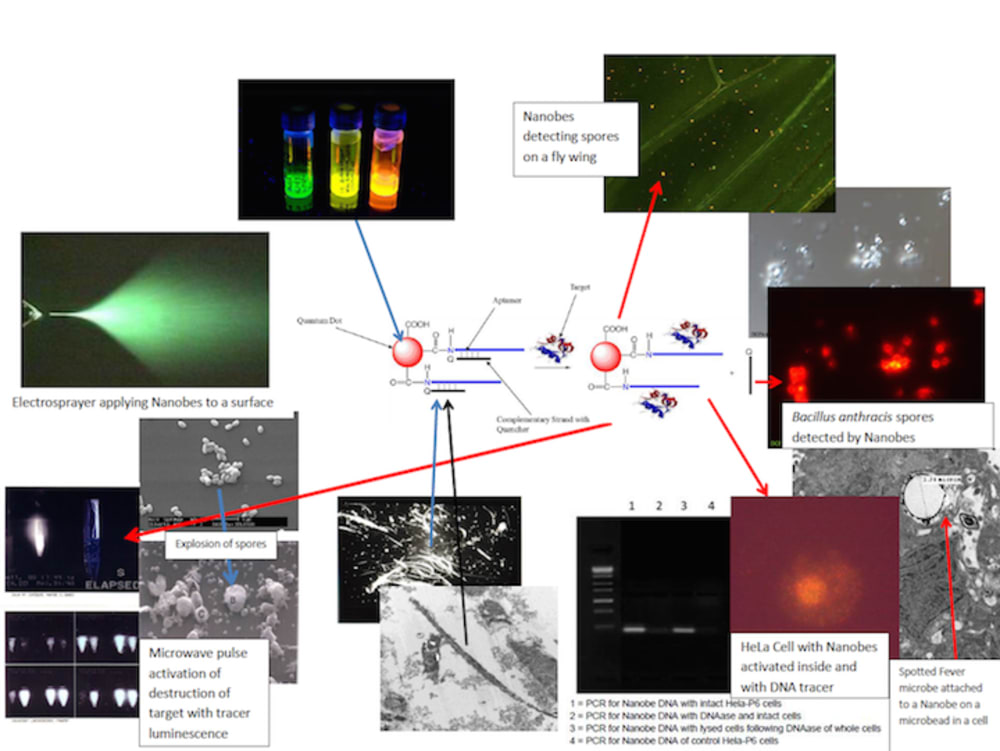
Nanobes are composites of aptamers, fluorescent dye or quantum dots, carbon nanotubes or quenching polymer (diazoluminomelanin) nanoparticles, and ferromagnetic metallic nanoparticles. The aptamer is a double-stranded DNA oligomer which dissociates upon contact with a specific target ligand (such as that found on the surface of a pathogen or toxin), separating one strand with the fluorescent material from the other DNA strand conjugated to the quencher of the fluorescence, the carbon nanotube or polymer. Upon separation, a fluorescent signal is produced which can be observed in bulk by a photodetector sensitive to the fluorescence or by a fluorescence microscope. The nanobes are “trapped” along with their attached target microbes or toxins by magnetic capture or separation in a microfluidics device, chromatographic device, in a microtiter plate format, free on contaminated surfaces, or in paper strips. The material can be observed, with the naked eye, with as little equipment as a magnet to trap the particles on absorbent surfaces and illumination with a UV flashlight for fluorescence.
More sophisticated microfluidics devices have already been built and demonstrated for this detection of positives, and technical information and patents can be provided. A microfluidics chamber in a current device built by Charles River has demonstrated that this detection can be carried out in a multichannel chamber which isolates the pathogen but retains its viability for further culture and analysis such as DNA/RNA isolation, PCR, and sequencing of its genome, or subunits of that genome, for confirmation of diagnosis and trace-back. Such confinement was designed for the safe handling of highly pathogenic infectious agents such as Ebola, anthrax spores, tularemia and Q fever bacteria (Francisella tularensis and Coxiella burnetii, respectively).
The ligand/anti-ligand pair can be replaced with either one or both of the DNA strands being exchanged for specific binding peptides, synthetic polymers, or antibody. DNA or RNA aptamers are preferred because of their stability, ease and cost effective synthetic selection and mass manufacture, and not requiring a cold chain. Other nanobes, used simultaneously, can transfect, on contact, the target bacteria and host cells, following the entry of the microbe (for viruses and rickettsia) with genes which express markers which demonstrate viability of the subject microbe. This property can be carried out for bacteria with minimal culture in media, which can be reconstituted with water, incorporated in the diagnostic chamber or test strips with a portable minimal incubator for use in the field.
For viruses and rickettsia, a mere positive in the initial test can be designed for a fragile target on the surface of the microbe which is absolutely required for infection (to test viability, infectivity, but not reproductive capability, in the presence of an antimicrobial; for these, it would require a host target cell, which would be problematic in the field, but the platform could be carried back to a forward lab for host cell culture). The chamber could be adapted to performing, in place, PCR on a captured virus or rickettsia which would also indicate viability.
Like this entry?
-
About the Entrant
- Name:Johnathan Kiel
- Type of entry:individual
- Patent status:patented